Matched Filter Chirp Search¶
Figure 10.27
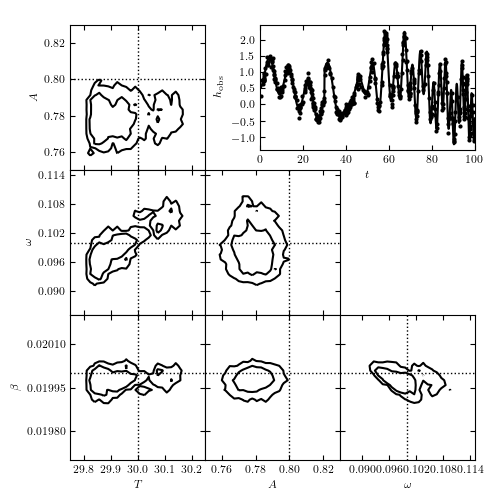
A ten-parameter chirp model (see eq. 10.87) fit to a time series. Seven of the parameters can be considered nuisance parameters, and we marginalize over them in the likelihood contours shown here.
# Author: Jake VanderPlas (adapted to PyMC3 by Brigitta Sipocz)
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
import pymc3 as pm
from astroML.plotting.mcmc import plot_mcmc
# ----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
# ----------------------------------------------------------------------
# Set up toy dataset
def chirp(t, T, A, phi, omega, beta):
"""chirp signal"""
mask = (t >= T)
signal = mask * (A * np.sin(phi + omega * (t - T) + beta * (t - T) ** 2))
return signal
def background(t, b0, b1, omega1, omega2):
"""background signal"""
return b0 + b1 * np.sin(omega1 * t) * np.sin(omega2 * t)
np.random.seed(0)
N = 500
T_true = 30
A_true = 0.8
phi_true = np.pi / 2
omega_true = 0.1
beta_true = 0.02
b0_true = 0.5
b1_true = 1.0
omega1_true = 0.3
omega2_true = 0.4
sigma = 0.1
t = 100 * np.random.random(N)
signal = chirp(t, T_true, A_true, phi_true, omega_true, beta_true)
bg = background(t, b0_true, b1_true, omega1_true, omega2_true)
y_true = signal + bg
y_obs = np.random.normal(y_true, sigma)
t_fit = np.linspace(0, 100, 1000)
y_fit = (chirp(t_fit, T_true, A_true, phi_true, omega_true, beta_true) +
background(t_fit, b0_true, b1_true, omega1_true, omega2_true))
# ----------------------------------------------------------------------
# Set up and run MCMC sampling
with pm.Model():
T = pm.Uniform('T', 0, 100, testval=T_true)
A = pm.Uniform('A', 0, 100, testval=A_true)
phi = pm.Uniform('phi', -np.pi, np.pi, testval=phi_true)
b0 = pm.Uniform('b0', 0, 100, testval=b0_true)
b1 = pm.Uniform('b1', 0, 100, testval=b1_true)
log_omega1 = pm.Uniform('log_omega1', -3, 0, testval=np.log(omega1_true))
log_omega2 = pm.Uniform('log_omega2', -3, 0, testval=np.log(omega2_true))
omega = pm.Uniform('omega', 0.001, 1, testval=omega_true)
beta = pm.Uniform('beta', 0.001, 1, testval=beta_true)
y = pm.Normal('y', mu=(chirp(t, T, A, phi, omega, beta)
+ background(t, b0, b1, np.exp(log_omega1), np.exp(log_omega2))),
tau=sigma ** -2, observed=y_obs)
step = pm.Metropolis()
traces = pm.sample(draws=5000, tune=2000, step=step)
labels = ['$T$', '$A$', r'$\omega$', r'$\beta$']
limits = [(29.75, 30.25), (0.75, 0.83), (0.085, 0.115), (0.0197, 0.0202)]
true = [T_true, A_true, omega_true, beta_true]
# ------------------------------------------------------------
# Plot results
fig = plt.figure(figsize=(5, 5))
# This function plots multiple panels with the traces
axes_list = plot_mcmc([traces[i] for i in ['T', 'A', 'omega', 'beta']],
labels=labels, limits=limits,
true_values=true, fig=fig,
bins=30, colors='k',
bounds=[0.14, 0.08, 0.95, 0.95])
for ax in axes_list:
for axis in [ax.xaxis, ax.yaxis]:
axis.set_major_locator(plt.MaxNLocator(5))
ax = fig.add_axes([0.52, 0.7, 0.43, 0.25])
ax.scatter(t, y_obs, s=9, lw=0, c='k')
ax.plot(t_fit, y_fit, '-k')
ax.set_xlim(0, 100)
ax.set_xlabel('$t$')
ax.set_ylabel(r'$h_{\rm obs}$')
plt.show()
