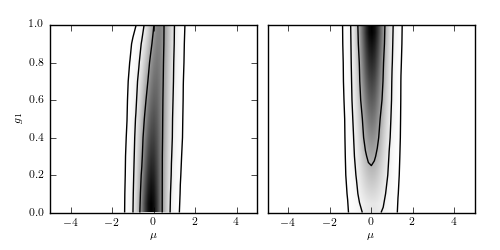
Plot the posterior of mu vs g1 with outliers¶
Figure 5.17
The marginal joint distribution between mu and g_i, as given by eq. 5.100.
The left panel shows a point identified as bad (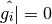 ),
while the right panel shows a point identified as good(
),
while the right panel shows a point identified as good(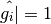 ).
).
# Author: Jake VanderPlas
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from scipy.stats import norm
from astroML.plotting.mcmc import convert_to_stdev
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
def p(mu, g1, xi, sigma1, sigma2):
"""Equation 5.97: marginalized likelihood over outliers"""
L = (g1 * norm.pdf(xi[0], mu, sigma1) +
(1 - g1) * norm.pdf(xi[0], mu, sigma2))
mu = mu.reshape(mu.shape + (1,))
g1 = g1.reshape(g1.shape + (1,))
return L * np.prod(norm.pdf(xi[1:], mu, sigma1)
+ norm.pdf(xi[1:], mu, sigma2), -1)
#------------------------------------------------------------
# Sample the points
np.random.seed(138)
N1 = 8
N2 = 2
sigma1 = 1
sigma2 = 3
sigmai = np.zeros(N1 + N2)
sigmai[N2:] = sigma1
sigmai[:N2] = sigma2
xi = np.random.normal(0, sigmai)
#------------------------------------------------------------
# Compute the marginalized posterior for the first and last point
mu = np.linspace(-5, 5, 71)
g1 = np.linspace(0, 1, 11)
L1 = p(mu[:, None], g1, xi, 1, 10)
L1 /= np.max(L1)
L2 = p(mu[:, None], g1, xi[::-1], 1, 10)
L2 /= np.max(L2)
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(5, 2.5))
fig.subplots_adjust(left=0.1, right=0.95, wspace=0.05,
bottom=0.15, top=0.9)
ax1 = fig.add_subplot(121)
ax1.imshow(L1.T, origin='lower', aspect='auto', cmap=plt.cm.binary,
extent=[mu[0], mu[-1], g1[0], g1[-1]])
ax1.contour(mu, g1, convert_to_stdev(np.log(L1).T),
levels=(0.683, 0.955, 0.997),
colors='k')
ax1.set_xlabel(r'$\mu$')
ax1.set_ylabel(r'$g_1$')
ax2 = fig.add_subplot(122)
ax2.imshow(L2.T, origin='lower', aspect='auto', cmap=plt.cm.binary,
extent=[mu[0], mu[-1], g1[0], g1[-1]])
ax2.contour(mu, g1, convert_to_stdev(np.log(L2).T),
levels=(0.683, 0.955, 0.997),
colors='k')
ax2.set_xlabel(r'$\mu$')
ax2.yaxis.set_major_locator(plt.NullLocator())
plt.show()
