Nonlinear cosmology fit to mu vs z¶
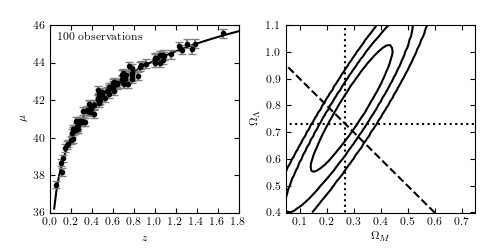
Figure 8.5
Cosmology fit to the standard cosmological integral. Errors in mu are a factor
of ten smaller than for the sample used in figure 8.2. Contours are 1-sigma,
2-sigma, and 3-sigma for the posterior (uniform prior in 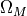 and
and
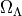 ). The dashed line shows flat cosmology. The dotted lines
show the input values.
). The dashed line shows flat cosmology. The dotted lines
show the input values.
@pickle_results: computing results and saving to 'mu_z_nonlinear.pkl'
# Author: Jake VanderPlas
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
from __future__ import print_function, division
import numpy as np
from matplotlib import pyplot as plt
from astropy.cosmology import LambdaCDM
from astroML.datasets import generate_mu_z
from astroML.plotting.mcmc import convert_to_stdev
from astroML.utils.decorators import pickle_results
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
#------------------------------------------------------------
# Generate the data
z_sample, mu_sample, dmu = generate_mu_z(100, z0=0.3,
dmu_0=0.05, dmu_1=0.004,
random_state=1)
#------------------------------------------------------------
# define a log likelihood in terms of the parameters
# beta = [omegaM, omegaL]
def compute_logL(beta):
cosmo = LambdaCDM(H0=71, Om0=beta[0], Ode0=beta[1], Tcmb0=0)
mu_pred = cosmo.distmod(z_sample).value
return - np.sum(0.5 * ((mu_sample - mu_pred) / dmu) ** 2)
#------------------------------------------------------------
# Define a function to compute (and save to file) the log-likelihood
@pickle_results('mu_z_nonlinear.pkl')
def compute_mu_z_nonlinear(Nbins=50):
omegaM = np.linspace(0.05, 0.75, Nbins)
omegaL = np.linspace(0.4, 1.1, Nbins)
logL = np.empty((Nbins, Nbins))
for i in range(len(omegaM)):
for j in range(len(omegaL)):
logL[i, j] = compute_logL([omegaM[i], omegaL[j]])
return omegaM, omegaL, logL
omegaM, omegaL, res = compute_mu_z_nonlinear()
res -= np.max(res)
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(5, 2.5))
fig.subplots_adjust(left=0.1, right=0.95, wspace=0.25,
bottom=0.15, top=0.9)
# left plot: the data and best-fit
ax = fig.add_subplot(121)
whr = np.where(res == np.max(res))
omegaM_best = omegaM[whr[0][0]]
omegaL_best = omegaL[whr[1][0]]
cosmo = LambdaCDM(H0=71, Om0=omegaM_best, Ode0=omegaL_best, Tcmb0=0)
z_fit = np.linspace(0.04, 2, 100)
mu_fit = cosmo.distmod(z_fit).value
ax.plot(z_fit, mu_fit, '-k')
ax.errorbar(z_sample, mu_sample, dmu, fmt='.k', ecolor='gray')
ax.set_xlim(0, 1.8)
ax.set_ylim(36, 46)
ax.set_xlabel('$z$')
ax.set_ylabel(r'$\mu$')
ax.text(0.04, 0.96, "%i observations" % len(z_sample),
ha='left', va='top', transform=ax.transAxes)
# right plot: the likelihood
ax = fig.add_subplot(122)
ax.contour(omegaM, omegaL, convert_to_stdev(res.T),
levels=(0.683, 0.955, 0.997),
colors='k')
ax.plot([0, 1], [1, 0], '--k')
ax.plot([0, 1], [0.73, 0.73], ':k')
ax.plot([0.27, 0.27], [0, 2], ':k')
ax.set_xlim(0.05, 0.75)
ax.set_ylim(0.4, 1.1)
ax.set_xlabel(r'$\Omega_M$')
ax.set_ylabel(r'$\Omega_\Lambda$')
plt.show()
