EM example: Gaussian Mixture Models¶
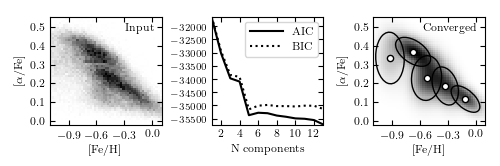
Figure 6.6
A two-dimensional mixture of Gaussians for the stellar metallicity data. The left panel shows the number density of stars as a function of two measures of their chemical composition: metallicity ([Fe/H]) and alpha-element abundance ([alpha/Fe]). The right panel shows the density estimated using mixtures of Gaussians together with the positions and covariances (2-sigma levels) of those Gaussians. The center panel compares the information criteria AIC and BIC (see Sections 4.3.2 and 5.4.3).
@pickle_results: computing results and saving to 'GMM_metallicity.pkl'
1
2
3
4
5
6
7
8
9
10
11
12
13
best fit converged: True
BIC: n_components = 5
# Author: Jake VanderPlas
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
from __future__ import print_function
import numpy as np
from matplotlib import pyplot as plt
from sklearn.mixture import GaussianMixture
from astroML.datasets import fetch_sdss_sspp
from astroML.utils.decorators import pickle_results
from astroML.plotting.tools import draw_ellipse
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
#------------------------------------------------------------
# Get the Segue Stellar Parameters Pipeline data
data = fetch_sdss_sspp(cleaned=True)
X = np.vstack([data['FeH'], data['alphFe']]).T
# truncate dataset for speed
X = X[::5]
#------------------------------------------------------------
# Compute GaussianMixture models & AIC/BIC
N = np.arange(1, 14)
@pickle_results("GMM_metallicity.pkl")
def compute_GaussianMixture(N, covariance_type='full', max_iter=1000):
models = [None for n in N]
for i in range(len(N)):
print(N[i])
models[i] = GaussianMixture(n_components=N[i], max_iter=max_iter,
covariance_type=covariance_type)
models[i].fit(X)
return models
models = compute_GaussianMixture(N)
AIC = [m.aic(X) for m in models]
BIC = [m.bic(X) for m in models]
i_best = np.argmin(BIC)
gmm_best = models[i_best]
print("best fit converged:", gmm_best.converged_)
print("BIC: n_components = %i" % N[i_best])
#------------------------------------------------------------
# compute 2D density
FeH_bins = 51
alphFe_bins = 51
H, FeH_bins, alphFe_bins = np.histogram2d(data['FeH'], data['alphFe'],
(FeH_bins, alphFe_bins))
Xgrid = np.array(list(map(np.ravel,
np.meshgrid(0.5 * (FeH_bins[:-1]
+ FeH_bins[1:]),
0.5 * (alphFe_bins[:-1]
+ alphFe_bins[1:]))))).T
log_dens = gmm_best.score_samples(Xgrid).reshape((51, 51))
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(5, 1.66))
fig.subplots_adjust(wspace=0.45,
bottom=0.25, top=0.9,
left=0.1, right=0.97)
# plot density
ax = fig.add_subplot(131)
ax.imshow(H.T, origin='lower', interpolation='nearest', aspect='auto',
extent=[FeH_bins[0], FeH_bins[-1],
alphFe_bins[0], alphFe_bins[-1]],
cmap=plt.cm.binary)
ax.set_xlabel(r'$\rm [Fe/H]$')
ax.set_ylabel(r'$\rm [\alpha/Fe]$')
ax.xaxis.set_major_locator(plt.MultipleLocator(0.3))
ax.set_xlim(-1.101, 0.101)
ax.text(0.93, 0.93, "Input",
va='top', ha='right', transform=ax.transAxes)
# plot AIC/BIC
ax = fig.add_subplot(132)
ax.plot(N, AIC, '-k', label='AIC')
ax.plot(N, BIC, ':k', label='BIC')
ax.legend(loc=1)
ax.set_xlabel('N components')
plt.setp(ax.get_yticklabels(), fontsize=7)
# plot best configurations for AIC and BIC
ax = fig.add_subplot(133)
ax.imshow(np.exp(log_dens),
origin='lower', interpolation='nearest', aspect='auto',
extent=[FeH_bins[0], FeH_bins[-1],
alphFe_bins[0], alphFe_bins[-1]],
cmap=plt.cm.binary)
ax.scatter(gmm_best.means_[:, 0], gmm_best.means_[:, 1], c='w')
for mu, C, w in zip(gmm_best.means_, gmm_best.covariances_, gmm_best.weights_):
draw_ellipse(mu, C, scales=[1.5], ax=ax, fc='none', ec='k')
ax.text(0.93, 0.93, "Converged",
va='top', ha='right', transform=ax.transAxes)
ax.set_xlim(-1.101, 0.101)
ax.set_ylim(alphFe_bins[0], alphFe_bins[-1])
ax.xaxis.set_major_locator(plt.MultipleLocator(0.3))
ax.set_xlabel(r'$\rm [Fe/H]$')
ax.set_ylabel(r'$\rm [\alpha/Fe]$')
plt.show()
